Molecular Biology
Molecular Genetics
Molecular Genetics
This site deals with Molecular Genetics, particularly in relation to genetic mechanisms of biological evolution. The site is searchable using the "Search this blog" box at top left, though Google is slow to update the search. Blue terms link to explanatory items so that you may navigate through items providing more detail, and the site is hyperlinked to the Companion Sites listed in the sidebar [indices]. The Molecular Genetics Overview or the SITE MAP lead into the items. Use the "back" function to return to each departure item.
|
0 Guide-Glossary


Molecular Genetics Overview
Molecular genetics is the study of molecules and mechanisms involved in genetic inheritance. Archival information molecules are long polymers of deoxyribonucleic acid (DNA) comprising bases in specific sequence. The bases adenine (A), thymine (T), cytosine (C), and guanine (G), function as codon triplets – sequences of three bases that code for specific amino acids or for translation initiation (start codons) or termination (stop codons). Uracil is substituted for thymine in RNA.
The segments of DNA that contain protein-coding instructions are called genes, and these gene sequences comprise a portion of the total genome of a cell. The genome includes both the genes (coding-sequences, domains) and the non-coding sequences – both exons, which include open reading frames, and introns.
Because the 64 possible combinations of GATC code for only the 20 amino acids commonly found in proteins, the code is 'degenerate' (redundant) with more than one triplet combination coding for each amino acid. (This code reduncancy provides hereditary stability by reducing mutation mistakes.) The double helix of DNA comprises paired nucleotide strands with bases hydrogen bonded to complementary bases in the adjacent chain. Adenine pairs with thymine or uracil (A-TU), and cytosine pairs with guanine (CG).
During cellular reproduction, strands of archival DNA are copied or replicated. Transcription is the first step in gene expression – DNA instructions are converted into mRNA codons, rRNAs, miRNAs, and tRNAs. Coding instructions of nucleotide sequences in archival DNA, which have been transcribed and processed into mRNAs are translated into polypeptides and proteins at cytoplasmic ribosomes. Translation is the ultimate step in gene expression, in which archival genetic instructions are converted into specified sequences of amino acids in peptides, polypeptides, and proteins.
In prokaryotic cells – without a nuclear membrane – translation into polypeptides and proteins may begin prior to termination of transcription. The molecular genetics of eukaryotic cells is more complicated than that of prokaryotes. Various molecules of ribonucleic acid (RNA) participate in the transcription of the DNA code into processed mRNA in a series of RNA processing stages including capping, polyadenylation, and pre-mRNA splicing.
Following pre-mRNA processing, RNAs undergo extranuclear transfer. Mature RNAs may undergo post-transcriptional modulation (via miRNAs) before translation of the archival DNA instructions into specific sequences of amino acids in the polypeptides and proteins that participate in cellular function and structure. Transfer RNAs (tRNA) deliver specific amino acids to the cytoplasmic ribosomes along the rough endoplasmic reticulum. Ribosomal RNAs participate in assembly of polypeptides and proteins at ribosomes. Here RNAs serve as ribozymes – non-protein enzymes.
A number of processes are involved in control of cellular function through the maintenance of accuracy of genetic inheritance – damage to DNA is repaired, and faulty RNA is destroyed.
DNA damage may result from replication errors, incorporation of mismatched nucleotides (substitution errors – transitions and transversions), damage by oxygen radicals, hydroxyl radicals, ionizing or ultraviolet radiation, toxins, alkylating agents, and chemotherapy agents. A number of vital mechanisms repair DNA damage to bases (including C to T, C to U, and T U mismatch) and to strands, including double strand breaks. All organisms, prokaryotic and eukaryotic, utilize at least three enzymatic excision-repair mechanisms for damaged bases: base excision repair, mismatch repair, and nucleotide excision repair.
Given the importance of mRNA as an information-carrying molecule, faulty pre-mRNAs and mRNAs must be eliminated – they are destroyed by nonsense-mediated decay or nonstop decay:
1. A pre-mRNA made from a mutant gene usually has an exon junction complex (EJC) in the wrong position. This error activates nonsense-mediated decay (NMD) and destroys the pre-mRNA before it can be used to make flawed proteins. There are at least two kinds of NMD: one requires the protein UPF2 and the other does not.
2. Nonstop decay is mRNA turnover mechanism that has none of the properties of normal mRNA turnover or of NMD. A multi-enzyme complex called the exosome is important for nonstop decay. The exosome is the site for binding of a specific adapter protein called Ski7p. Nonstop decay shares none of the enzymes required for nonsense-mediated decay.
Just as cells repair DNA, they must also maintain the proteome by managing damaged proteins. Heat stress denaturates proteins, causing weakening of polar bonds and exposure of hydrophobic groups. The cellular stress response (heat-shock response) protects organisms from damage resulting from environmental stressors such as heat, UV light, trace metals, and xenobiotics. Stress genes are activated to rapidly synthesize stress proteins, which are highly conserved in biological evolution and play similar roles in organisms from bacteria to humans. Normally, several constitutive stress proteins are present at low levels to function as molecular chaperones, so as to facilitate folding, assembly, and distribution of newly synthesized proteins. For the environmentally stressed cell, stress proteins protect and repair vulnerable protein targets, and play a role in the lysosomal and ubiquitin protein degradation pathways (for removal of unsalvageable proteins). Thus, the cellular stress response performs orchestrated induction of key proteins necessary for cellular protein repair and degradation systems. | 0 Guide-Glossary

The segments of DNA that contain protein-coding instructions are called genes, and these gene sequences comprise a portion of the total genome of a cell. The genome includes both the genes (coding-sequences, domains) and the non-coding sequences – both exons, which include open reading frames, and introns.
Because the 64 possible combinations of GATC code for only the 20 amino acids commonly found in proteins, the code is 'degenerate' (redundant) with more than one triplet combination coding for each amino acid. (This code reduncancy provides hereditary stability by reducing mutation mistakes.) The double helix of DNA comprises paired nucleotide strands with bases hydrogen bonded to complementary bases in the adjacent chain. Adenine pairs with thymine or uracil (A-TU), and cytosine pairs with guanine (CG).
During cellular reproduction, strands of archival DNA are copied or replicated. Transcription is the first step in gene expression – DNA instructions are converted into mRNA codons, rRNAs, miRNAs, and tRNAs. Coding instructions of nucleotide sequences in archival DNA, which have been transcribed and processed into mRNAs are translated into polypeptides and proteins at cytoplasmic ribosomes. Translation is the ultimate step in gene expression, in which archival genetic instructions are converted into specified sequences of amino acids in peptides, polypeptides, and proteins.
In prokaryotic cells – without a nuclear membrane – translation into polypeptides and proteins may begin prior to termination of transcription. The molecular genetics of eukaryotic cells is more complicated than that of prokaryotes. Various molecules of ribonucleic acid (RNA) participate in the transcription of the DNA code into processed mRNA in a series of RNA processing stages including capping, polyadenylation, and pre-mRNA splicing.
Following pre-mRNA processing, RNAs undergo extranuclear transfer. Mature RNAs may undergo post-transcriptional modulation (via miRNAs) before translation of the archival DNA instructions into specific sequences of amino acids in the polypeptides and proteins that participate in cellular function and structure. Transfer RNAs (tRNA) deliver specific amino acids to the cytoplasmic ribosomes along the rough endoplasmic reticulum. Ribosomal RNAs participate in assembly of polypeptides and proteins at ribosomes. Here RNAs serve as ribozymes – non-protein enzymes.
A number of processes are involved in control of cellular function through the maintenance of accuracy of genetic inheritance – damage to DNA is repaired, and faulty RNA is destroyed.
DNA damage may result from replication errors, incorporation of mismatched nucleotides (substitution errors – transitions and transversions), damage by oxygen radicals, hydroxyl radicals, ionizing or ultraviolet radiation, toxins, alkylating agents, and chemotherapy agents. A number of vital mechanisms repair DNA damage to bases (including C to T, C to U, and T U mismatch) and to strands, including double strand breaks. All organisms, prokaryotic and eukaryotic, utilize at least three enzymatic excision-repair mechanisms for damaged bases: base excision repair, mismatch repair, and nucleotide excision repair.
Given the importance of mRNA as an information-carrying molecule, faulty pre-mRNAs and mRNAs must be eliminated – they are destroyed by nonsense-mediated decay or nonstop decay:
1. A pre-mRNA made from a mutant gene usually has an exon junction complex (EJC) in the wrong position. This error activates nonsense-mediated decay (NMD) and destroys the pre-mRNA before it can be used to make flawed proteins. There are at least two kinds of NMD: one requires the protein UPF2 and the other does not.
2. Nonstop decay is mRNA turnover mechanism that has none of the properties of normal mRNA turnover or of NMD. A multi-enzyme complex called the exosome is important for nonstop decay. The exosome is the site for binding of a specific adapter protein called Ski7p. Nonstop decay shares none of the enzymes required for nonsense-mediated decay.
Just as cells repair DNA, they must also maintain the proteome by managing damaged proteins. Heat stress denaturates proteins, causing weakening of polar bonds and exposure of hydrophobic groups. The cellular stress response (heat-shock response) protects organisms from damage resulting from environmental stressors such as heat, UV light, trace metals, and xenobiotics. Stress genes are activated to rapidly synthesize stress proteins, which are highly conserved in biological evolution and play similar roles in organisms from bacteria to humans. Normally, several constitutive stress proteins are present at low levels to function as molecular chaperones, so as to facilitate folding, assembly, and distribution of newly synthesized proteins. For the environmentally stressed cell, stress proteins protect and repair vulnerable protein targets, and play a role in the lysosomal and ubiquitin protein degradation pathways (for removal of unsalvageable proteins). Thus, the cellular stress response performs orchestrated induction of key proteins necessary for cellular protein repair and degradation systems. | 0 Guide-Glossary
alternative splicing
Alternative splicing is a carefully regulated, variable adaptation of the constitutive RNA modification process of pre-mRNA splicing. Alternative splicing is a form of epigenetic mechanism that enables a single gene to give rise to multiple, differentially spliced versions of a protein, increasing complexity without a change in the genome. ... MORE
Biochemistry Overview : Molecular Genetics Overview : SITE MAP : HOME | 0 Guide-Glossary

Biochemistry Overview : Molecular Genetics Overview : SITE MAP : HOME | 0 Guide-Glossary
transcription
In transcription, an RNA polymerase enzyme (RNAp, or pol III in eukaryotes) directs generation of a complementary strand of mRNA from DNA. The mechanism of alternative splicing enables to production of different mature mRNA molecules, depending on what sequences are treated as introns and what remain as exons.
Transcription involves 3 phases: initiation, elongation, and termination.
1. RNA polymerase II initiates transcription at the first nucleotide of the first exon of a gene.
2. The 5 end of the nascent RNA is capped with 7-methylguanylate (capping).
3. Transcription by RNA polymerase II terminates at any one of multiple termination sites downstream from the poly(A) site, which is located at the 3 end of the final exon.
4. After the primary transcript is cleaved at the poly(A) site,
5. A string of adenine (A) residues is added (polyadenylation). The poly(A) tail contains ≈250 A residues in mammals, ≈150 in insects, and ≈100 in yeasts.
In more detail: RNA polymerase binds to the promoter region of one strand of DNA (5’ end), and the DNA double helix is un-zipped into single strands. First, RNA polymerase requires a number of general transcription factors (called TFIIA, TFIIB, etc.). The promoter contains a DNA sequence called the TATA box, which is located 25 nucleotides away from the site of initiation of transcription. The TATA box is recognized and bound by transcription factor TFIID, which then enables the adjacent binding of TFIIB. The rest of the general transcription factors plus the RNA polymerase assemble at the promoter. diagram - initiation of transcription.
The RNAp enzyme moves toward the 3’ end, connecting complementary bases into an elongating chain of RNA nucleotides. At termination, the transcribed mRNA molecule is released from the DNA strand. In prokaryotic cells – without a nuclear membrane – translation may begin prior to termination. In eukaryotic cells – with a nuclear membrane – the processed mRNA moves through the nuclear pores into the cytoplasm, where ribosomes on the rough endoplasmic reticulum translate the mRNA code into a peptide or a protein. Epigenetic, alternative splicing mechanisms can edit the mRNA prior to its translation into protein.
Capping of the 5’ end on the pre-mRNA with 7-methylguanylate occurs soon after initiation of transcription, and the 5 cap is retained in mature mRNAs.
Cleavage, pre-mRNA splicing, and polyadenylation usually follow termination of transcription of short primary transcripts with few introns. However, introns often are spliced out of the nascent RNA before transcription of the gene is complete for large genes with multiple introns.
It was believed that most genes in higher eukaryotes are regulated by controlling their transcription. However, it is increasingly recognized that epigenetic mechanisms (such as alternative splicing) are important in generating many proteins from a single gene, accounting for the Human Genome Project’s discovery that a mere 30,000 genes code for about 100,000 proteins.
animation - start of transcription : animation - life cycle of an mRNA : animation ~ alternative splicing More at NCBI Molecular Cell Biology - Transcription Initiation Complex : SUMMARY transcription initiation (NCBI MCB) : Processes of Transcription : Wikipedia : Central Dogma :
Biochemistry Overview : Molecular Genetics Overview : SITE MAP : HOME | 0 Guide-Glossary

Transcription involves 3 phases: initiation, elongation, and termination.
1. RNA polymerase II initiates transcription at the first nucleotide of the first exon of a gene.
2. The 5 end of the nascent RNA is capped with 7-methylguanylate (capping).
3. Transcription by RNA polymerase II terminates at any one of multiple termination sites downstream from the poly(A) site, which is located at the 3 end of the final exon.
4. After the primary transcript is cleaved at the poly(A) site,
5. A string of adenine (A) residues is added (polyadenylation). The poly(A) tail contains ≈250 A residues in mammals, ≈150 in insects, and ≈100 in yeasts.
In more detail: RNA polymerase binds to the promoter region of one strand of DNA (5’ end), and the DNA double helix is un-zipped into single strands. First, RNA polymerase requires a number of general transcription factors (called TFIIA, TFIIB, etc.). The promoter contains a DNA sequence called the TATA box, which is located 25 nucleotides away from the site of initiation of transcription. The TATA box is recognized and bound by transcription factor TFIID, which then enables the adjacent binding of TFIIB. The rest of the general transcription factors plus the RNA polymerase assemble at the promoter. diagram - initiation of transcription.
The RNAp enzyme moves toward the 3’ end, connecting complementary bases into an elongating chain of RNA nucleotides. At termination, the transcribed mRNA molecule is released from the DNA strand. In prokaryotic cells – without a nuclear membrane – translation may begin prior to termination. In eukaryotic cells – with a nuclear membrane – the processed mRNA moves through the nuclear pores into the cytoplasm, where ribosomes on the rough endoplasmic reticulum translate the mRNA code into a peptide or a protein. Epigenetic, alternative splicing mechanisms can edit the mRNA prior to its translation into protein.
Capping of the 5’ end on the pre-mRNA with 7-methylguanylate occurs soon after initiation of transcription, and the 5 cap is retained in mature mRNAs.
Cleavage, pre-mRNA splicing, and polyadenylation usually follow termination of transcription of short primary transcripts with few introns. However, introns often are spliced out of the nascent RNA before transcription of the gene is complete for large genes with multiple introns.
It was believed that most genes in higher eukaryotes are regulated by controlling their transcription. However, it is increasingly recognized that epigenetic mechanisms (such as alternative splicing) are important in generating many proteins from a single gene, accounting for the Human Genome Project’s discovery that a mere 30,000 genes code for about 100,000 proteins.
animation - start of transcription : animation - life cycle of an mRNA : animation ~ alternative splicing More at NCBI Molecular Cell Biology - Transcription Initiation Complex : SUMMARY transcription initiation (NCBI MCB) : Processes of Transcription : Wikipedia : Central Dogma :
Biochemistry Overview : Molecular Genetics Overview : SITE MAP : HOME | 0 Guide-Glossary
base excision repair
Base excision repair (BER) describes one form of excision repair in which damaged bases or incorrect bases are excised and replaced through specific enzymes that differ between species. However, the biochemical processes involved in BER are equivalent across species, so bacterial DNA repair functions can operate in eukaryotic cells, and vice versa. Damage is typically the result of deamination, alkylation, hydroxylation, or attack by an oxygen radical, while the incorrect base can be uracil substituted for thymine. Oxidative DNA lesions induced by oxygen free radicals such as superoxide and hydroxyl radicals appear to be repaired predominantly by base excision repair mechanisms. Further, BER is the major DNA repair system involved in removal of various oxidative DNA lesions induced by ionizing radiation - these include abasic sites and modified DNA base and sugar residues.
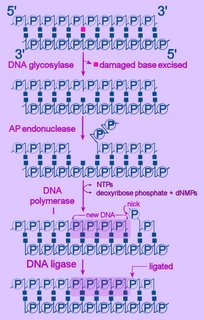 Left - diagram of base excision repair - click to enlarge image.
Left - diagram of base excision repair - click to enlarge image.
NTPs = ribonucleoside triphosphates
dNMP = deoxyribonucleoside monophosphate
First, the altered base is excised by a specific DNA glycosylase, which breaks the beta N-glycosidic bond and creates an AP, or abasic site. This site is identical to that generated by spontaneous depyrimidination or depurination. Six DNA glycosylases have been identified in humans – each excises an overlapping subset of either spontaneously formed (such as hypoxanthine), oxidized (such as 8-oxo-7,8-dihydroguanine), alkylated (such as 3-methyladenine), or mismatched (for example, T:G) bases.
Next, the terminal sugar-phosphate is removed by an AP endonuclease (Ape1), leaving a 3’-OH terminal and an abnormal 5'-abasic terminus. The resulting gap is refilled by the 5’-deoxyribose-phosphodiesterase action of a DNA polymerase I (DNA polymerase beta in mammals), then the strands are re-ligated by DNA Ligase I or a complex of XRCC1 and LigIII.
An alternative BER pathway corrects errors involving more than one nucleotide. The Fen1 protein excises the long-patch structure that is produced by DNA polymerase strand displacement. This "long-patch" repair process is divided into two subpathways: a PCNA-stimulated, Pol-beta-directed pathway and a PCNA-dependent, Pol-delta/epsilon -directed pathway.
link to table - human DNA repair genes : diagram>BER
Biochemistry Overview : Molecular Genetics Overview : SITE MAP : HOME | 0 Guide-Glossary

 Left - diagram of base excision repair - click to enlarge image.
Left - diagram of base excision repair - click to enlarge image.NTPs = ribonucleoside triphosphates
dNMP = deoxyribonucleoside monophosphate
First, the altered base is excised by a specific DNA glycosylase, which breaks the beta N-glycosidic bond and creates an AP, or abasic site. This site is identical to that generated by spontaneous depyrimidination or depurination. Six DNA glycosylases have been identified in humans – each excises an overlapping subset of either spontaneously formed (such as hypoxanthine), oxidized (such as 8-oxo-7,8-dihydroguanine), alkylated (such as 3-methyladenine), or mismatched (for example, T:G) bases.
Next, the terminal sugar-phosphate is removed by an AP endonuclease (Ape1), leaving a 3’-OH terminal and an abnormal 5'-abasic terminus. The resulting gap is refilled by the 5’-deoxyribose-phosphodiesterase action of a DNA polymerase I (DNA polymerase beta in mammals), then the strands are re-ligated by DNA Ligase I or a complex of XRCC1 and LigIII.
An alternative BER pathway corrects errors involving more than one nucleotide. The Fen1 protein excises the long-patch structure that is produced by DNA polymerase strand displacement. This "long-patch" repair process is divided into two subpathways: a PCNA-stimulated, Pol-beta-directed pathway and a PCNA-dependent, Pol-delta/epsilon -directed pathway.
link to table - human DNA repair genes : diagram>BER
Biochemistry Overview : Molecular Genetics Overview : SITE MAP : HOME | 0 Guide-Glossary
cis versus trans-acting factors
Most often, signal elements act only on the intramolecular nucleotide sequence to which they are attached, and they are said to act "in cis". Intron removal in eukaryotes involves cis-splicing. Interaction with signal factors — usually protein molecules — turns signal elements on or off. When protein factors are free to diffuse within the cell they can act on target elements that may not be derived from the same genome segment. Protein factors capable of acting upon other intermolecular genome segments are called "trans-acting factors".
One form of trans-splicing is the 'spliced leader' type, which is primarily found in protozoans (e.g. trypanosomes) and in lower invertebrates such as nematodes. This results in the addition of a capped, noncoding, spliced leader sequence to the 5' end of mRNAs.
Another form of trans-splicing is the 'discontinuous group II intron' type that occurs in plant/algal chloroplasts and plant mitochondria. This results in the joining of two independently transcribed coding sequences. Both spliced-leader and discontinuous group II intron trans-splicing are mechanistically similar to conventional nuclear pre-mRNA cis-splicing. Trans-splicing also occurs in mammalian cells, just as cis-splicing occurs in trypanosomes. It has been suggested that both trans- and cis-splicing are ancient acquisitions of the eukaryotic cell. (Abstract)
Biochemistry Overview : Molecular Genetics Overview : SITE MAP : HOME | 0 Guide-Glossary

One form of trans-splicing is the 'spliced leader' type, which is primarily found in protozoans (e.g. trypanosomes) and in lower invertebrates such as nematodes. This results in the addition of a capped, noncoding, spliced leader sequence to the 5' end of mRNAs.
Another form of trans-splicing is the 'discontinuous group II intron' type that occurs in plant/algal chloroplasts and plant mitochondria. This results in the joining of two independently transcribed coding sequences. Both spliced-leader and discontinuous group II intron trans-splicing are mechanistically similar to conventional nuclear pre-mRNA cis-splicing. Trans-splicing also occurs in mammalian cells, just as cis-splicing occurs in trypanosomes. It has been suggested that both trans- and cis-splicing are ancient acquisitions of the eukaryotic cell. (Abstract)
Biochemistry Overview : Molecular Genetics Overview : SITE MAP : HOME | 0 Guide-Glossary
capping
Capping is a form of RNA processing in which the 5’ end of the nascent pre-mRNA is capped with a 7-methyl guanosine nucleotide, 7-methylguanylate. Capping occurs shortly after initiation of transcription.
The 5' cap is retained in mature mRNAs. Capping is required to protect the RNA transcript from degradation. It plays an important role in mRNA transport to the cytoplasm and in the initiation of protein synthesis (translation) .
life cycle of an mRNA ~ click on Quicktime QNCBI Molecular Cell Biology Post-transcriptional Processing of RNAs
Biochemistry Overview : Molecular Genetics Overview : SITE MAP : HOME | 0 Guide-Glossary

The 5' cap is retained in mature mRNAs. Capping is required to protect the RNA transcript from degradation. It plays an important role in mRNA transport to the cytoplasm and in the initiation of protein synthesis (translation) .
life cycle of an mRNA ~ click on Quicktime QNCBI Molecular Cell Biology Post-transcriptional Processing of RNAs
Biochemistry Overview : Molecular Genetics Overview : SITE MAP : HOME | 0 Guide-Glossary
codon
Triplets of DNA nucleotides arranged in sequence along DNA are transcribed into the codons of mRNA strands.
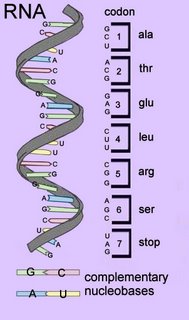 In the centre of the DNA double helix each nucleobase (base) is hydrogen bonded to its complementary base – because of steric (size) constraints, purines bases pair with pyrimidine bases. A with T, and C with G.
In the centre of the DNA double helix each nucleobase (base) is hydrogen bonded to its complementary base – because of steric (size) constraints, purines bases pair with pyrimidine bases. A with T, and C with G.
RNA molecules are created by transcription from the archival DNA template. Like replication, transcription proceeds 3' to 5' on the template strand and 5' to 3' on the RNA strand. Proteins are synthesized from the amino acid terminus to the carboxyl terminus by formation of peptide bonds between amino acids delivered by tRNA anticodons.
Because the nucleotide triplet sequences would produce different amino acids if ‘reading frames’ commenced at the wrong nucleotide, codons in genes are read in reading frames that commence at the start codon – usually the first AUG in the RNA sequence (ATG triplet for DNA). AUG also codes for the amino acid methionine.
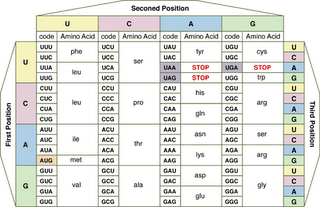 The RNA code is at left (click to enlarge). In accord with its origin early in evolution, the genetic code is almost universal – with only a few organisms employing variants.
The RNA code is at left (click to enlarge). In accord with its origin early in evolution, the genetic code is almost universal – with only a few organisms employing variants.
Each tri-nucleotide in the DNA sequence and transcribed mRNA codon signals for the insertion of one amino acid into a polymerizing peptide chain. The sequence of codons in genes determines the sequence of amino acids inserted into peptide, polypeptide and protein chains (primary structure).
Take an interactive peek at a sequence of DNA around the gene cyclooxygenase 2: PBS Cracking the Code of Life : Find on/off : Find start : Find stop : Find exons : Find introns : Find transposable elements : Find conserved DNA : Find single nucleotide polymorphisms : Find gene and scroll down:
Biochemistry Overview : Molecular Genetics Overview : SITE MAP : HOME | 0 Guide-Glossary

 In the centre of the DNA double helix each nucleobase (base) is hydrogen bonded to its complementary base – because of steric (size) constraints, purines bases pair with pyrimidine bases. A with T, and C with G.
In the centre of the DNA double helix each nucleobase (base) is hydrogen bonded to its complementary base – because of steric (size) constraints, purines bases pair with pyrimidine bases. A with T, and C with G.RNA molecules are created by transcription from the archival DNA template. Like replication, transcription proceeds 3' to 5' on the template strand and 5' to 3' on the RNA strand. Proteins are synthesized from the amino acid terminus to the carboxyl terminus by formation of peptide bonds between amino acids delivered by tRNA anticodons.
Because the nucleotide triplet sequences would produce different amino acids if ‘reading frames’ commenced at the wrong nucleotide, codons in genes are read in reading frames that commence at the start codon – usually the first AUG in the RNA sequence (ATG triplet for DNA). AUG also codes for the amino acid methionine.
 The RNA code is at left (click to enlarge). In accord with its origin early in evolution, the genetic code is almost universal – with only a few organisms employing variants.
The RNA code is at left (click to enlarge). In accord with its origin early in evolution, the genetic code is almost universal – with only a few organisms employing variants.Each tri-nucleotide in the DNA sequence and transcribed mRNA codon signals for the insertion of one amino acid into a polymerizing peptide chain. The sequence of codons in genes determines the sequence of amino acids inserted into peptide, polypeptide and protein chains (primary structure).
Take an interactive peek at a sequence of DNA around the gene cyclooxygenase 2: PBS Cracking the Code of Life : Find on/off : Find start : Find stop : Find exons : Find introns : Find transposable elements : Find conserved DNA : Find single nucleotide polymorphisms : Find gene and scroll down:
Biochemistry Overview : Molecular Genetics Overview : SITE MAP : HOME | 0 Guide-Glossary








































